nodes Package¶
nodes Package¶
nodes using the mdp-toolkit node interface
alignment Module¶
initialization - alignment of spike waveform sets
-
class
AlignmentNode(nchan=4, max_rep=32, max_tau=10, resample_factor=None, cut_down=True, dtype=<type 'numpy.float32'>, debug=False)[source]¶ Bases:
botmpy.nodes.base_nodes.ResetNodealigns a set of spikes on the mean waveform of the set
-
execute(x)¶ Process the data contained in x.
If the object is still in the training phase, the function stop_training will be called. x is a matrix having different variables on different columns and observations on the rows.
By default, subclasses should overwrite _execute to implement their execution phase. The docstring of the _execute method overwrites this docstring.
-
artifact_detector Module¶
detector nodes for capacitative artifacts in multichanneled data
These detectors find events and event epochs on potentially multichanneled data signal. Mostly, you will want to reset the internals of the detector after processing a chunk of data. There are different kinds of detectors, the common product of the detector is the discrete events or epochs in the data signal.
DATA_TYPE: fixed for float32 (single precision)
-
class
ArtifactDetectorNode(wsize_ms=15.0, psize_ms=(5.0, 10.0), wfunc=<function ones>, srate=32000.0, zcr_th=0.1, mindist_ms=10.0)[source]¶ Bases:
botmpy.nodes.spike_detection.ThresholdDetectorNodedetects artifacts by detecting zero-crossing frequencies
For a zero-mean gaussian process the the zero-crossing rate zcr is independent of its moments and approaches 0.5 as the integration window size approaches infinity:
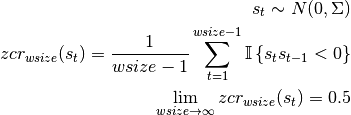
The capacitive artifacts seen in the Munk dataset have a significantly lower frequency, s.t. zcr decreases to 0.1 and below, for the integration window sizes relevant to our application. Detecting epochs where the zcr significantly deviates from the expectation, assuming a coloured Gaussian noise process, can thus lead be used for detection of artifact epochs.
The zero crossing rate (zcr) is given by the convolution of a moving average window (although this is configurable to use other weighting methods) with the XOR of the signbits of X(t) and X(t+1).
-
execute(x, *args, **kwargs)¶ calls self._apply_threshold() and return the events found
-
-
class
SpectrumArtifactDetector(wsize_ms=8.0, srate=32000.0, cutoff_hz=2000.0, nfft=512, en_func='max_normed', overlap=1, max_merge_dist=6, min_allowed_length=2, **kw)[source]¶ Bases:
botmpy.nodes.spike_detection.ThresholdDetectorNodedetects artifacts by identifying unwanted frequency packages in the spectrum of the signal
For a zero-mean gaussian process the the zero-crossing rate zcr is independent of its moments and approaches 0.5 as the integration window size approaches infinity:
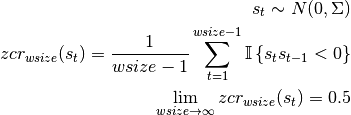
The capacitive artifacts seen in the Munk dataset have a significantly lower frequency, s.t. zcr decreases to 0.1 and below, for the integration window sizes relevant to our application. Detecting epochs where the zcr significantly deviates from the expectation, assuming a coloured Gaussian noise process, can thus lead be used for detection of artifact epochs.
The zero crossing rate (zcr) is given by the convolution of a moving average window (although this is configurable to use other weighting methods) with the XOR of the signbits of X(t) and X(t+1).
-
execute(x, *args, **kwargs)¶ calls self._apply_threshold() and return the events found
-
base_nodes Module¶
abstract base classes derived from MDP nodes
-
class
Node(input_dim=None, output_dim=None, dtype=None)¶ Bases:
future.types.newobject.newobjectA Node is the basic building block of an MDP application.
It represents a data processing element, like for example a learning algorithm, a data filter, or a visualization step. Each node can have one or more training phases, during which the internal structures are learned from training data (e.g. the weights of a neural network are adapted or the covariance matrix is estimated) and an execution phase, where new data can be processed forwards (by processing the data through the node) or backwards (by applying the inverse of the transformation computed by the node if defined).
Nodes have been designed to be applied to arbitrarily long sets of data: if the underlying algorithms supports it, the internal structures can be updated incrementally by sending multiple batches of data (this is equivalent to online learning if the chunks consists of single observations, or to batch learning if the whole data is sent in a single chunk). It is thus possible to perform computations on amounts of data that would not fit into memory or to generate data on-the-fly.
A Node also defines some utility methods, like for example copy and save, that return an exact copy of a node and save it in a file, respectively. Additional methods may be present, depending on the algorithm.
Node subclasses should take care of overwriting (if necessary) the functions is_trainable, _train, _stop_training, _execute, is_invertible, _inverse, _get_train_seq, and _get_supported_dtypes. If you need to overwrite the getters and setters of the node’s properties refer to the docstring of get_input_dim/set_input_dim, get_output_dim/set_output_dim, and get_dtype/set_dtype.
-
copy(protocol=None)¶ Return a deep copy of the node.
Parameters: protocol – the pickle protocol (deprecated).
-
dtype¶
-
execute(x, *args, **kwargs)¶ Process the data contained in x.
If the object is still in the training phase, the function stop_training will be called. x is a matrix having different variables on different columns and observations on the rows.
By default, subclasses should overwrite _execute to implement their execution phase. The docstring of the _execute method overwrites this docstring.
-
get_current_train_phase()¶ Return the index of the current training phase.
The training phases are defined in the list self._train_seq.
-
get_dtype()¶ Return dtype.
-
get_input_dim()¶ Return input dimensions.
-
get_output_dim()¶ Return output dimensions.
-
get_remaining_train_phase()¶ Return the number of training phases still to accomplish.
If the node is not trainable then return 0.
-
get_supported_dtypes()¶ Return dtypes supported by the node as a list of :numpy:`dtype` objects.
Note that subclasses should overwrite self._get_supported_dtypes when needed.
-
has_multiple_training_phases()¶ Return True if the node has multiple training phases.
-
input_dim¶ Input dimensions
-
inverse(y, *args, **kwargs)¶ Invert y.
If the node is invertible, compute the input
xsuch thaty = execute(x).By default, subclasses should overwrite _inverse to implement their inverse function. The docstring of the inverse method overwrites this docstring.
-
static
is_invertible()¶ Return True if the node can be inverted, False otherwise.
-
static
is_trainable()¶ Return True if the node can be trained, False otherwise.
-
is_training()¶ Return True if the node is in the training phase, False otherwise.
-
output_dim¶ Output dimensions
-
save(filename, protocol=-1)¶ Save a pickled serialization of the node to filename. If filename is None, return a string.
Note: the pickled Node is not guaranteed to be forwards or backwards compatible.
-
set_dtype(t)¶ Set internal structures’ dtype.
Perform sanity checks and then calls
self._set_dtype(n), which is responsible for setting the internal attributeself._dtype. Note that subclasses should overwrite self._set_dtype when needed.
-
set_input_dim(n)¶ Set input dimensions.
Perform sanity checks and then calls
self._set_input_dim(n), which is responsible for setting the internal attributeself._input_dim. Note that subclasses should overwrite self._set_input_dim when needed.
-
set_output_dim(n)¶ Set output dimensions.
Perform sanity checks and then calls
self._set_output_dim(n), which is responsible for setting the internal attributeself._output_dim. Note that subclasses should overwrite self._set_output_dim when needed.
-
stop_training(*args, **kwargs)¶ Stop the training phase.
By default, subclasses should overwrite _stop_training to implement this functionality. The docstring of the _stop_training method overwrites this docstring.
-
supported_dtypes¶ Supported dtypes
-
train(x, *args, **kwargs)¶ Update the internal structures according to the input data x.
x is a matrix having different variables on different columns and observations on the rows.
By default, subclasses should overwrite _train to implement their training phase. The docstring of the _train method overwrites this docstring.
Note: a subclass supporting multiple training phases should implement the same signature for all the training phases and document the meaning of the arguments in the _train method doc-string. Having consistent signatures is a requirement to use the node in a flow.
-
-
class
ResetNode(input_dim=None, output_dim=None, dtype=None)[source]¶ Bases:
botmpy.nodes.base_nodes.TrainingResetMixin,mdp.Node
-
class
TrainingResetMixin[source]¶ Bases:
objectallows
mdp.Nodeto reset to training stateThis is a mixin class for subclasses of
mdp.Node. To use it inherit frommdp.Nodeand put this mixin as the first superclass.node is a mdp.signal_node.Cumulator that can have its training phase reinitialised once a batch of cumulated data has been processed on. This is useful for online algorithms that derive parameters from the batch of data currently under consideration (Ex.: stochastic thresholding).
-
class
PCANode(input_dim=None, output_dim=None, dtype=None, svd=False, reduce=False, var_rel=1e-12, var_abs=1e-15, var_part=None)¶ Bases:
mdp.NodeFilter the input data through the most significatives of its principal components.
Internal variables of interest
self.avg- Mean of the input data (available after training).
self.v- Transposed of the projection matrix (available after training).
self.d- Variance corresponding to the PCA components (eigenvalues of the covariance matrix).
self.explained_variance- When output_dim has been specified as a fraction of the total variance, this is the fraction of the total variance that is actually explained.
More information about Principal Component Analysis, a.k.a. discrete Karhunen-Loeve transform can be found among others in I.T. Jolliffe, Principal Component Analysis, Springer-Verlag (1986).
-
execute(x, n=None)¶ Project the input on the first ‘n’ principal components. If ‘n’ is not set, use all available components.
-
get_explained_variance()¶ Return the fraction of the original variance that can be explained by self._output_dim PCA components. If for example output_dim has been set to 0.95, the explained variance could be something like 0.958... Note that if output_dim was explicitly set to be a fixed number of components, there is no way to calculate the explained variance.
-
get_projmatrix(transposed=1)¶ Return the projection matrix.
-
get_recmatrix(transposed=1)¶ Return the back-projection matrix (i.e. the reconstruction matrix).
-
inverse(y, n=None)¶ Project ‘y’ to the input space using the first ‘n’ components. If ‘n’ is not set, use all available components.
-
stop_training(debug=False)¶ Stop the training phase.
Keyword arguments:
- debug=True if stop_training fails because of singular cov
- matrices, the singular matrices itselves are stored in self.cov_mtx and self.dcov_mtx to be examined.
-
train(x)¶ Update the internal structures according to the input data x.
x is a matrix having different variables on different columns and observations on the rows.
By default, subclasses should overwrite _train to implement their training phase. The docstring of the _train method overwrites this docstring.
Note: a subclass supporting multiple training phases should implement the same signature for all the training phases and document the meaning of the arguments in the _train method doc-string. Having consistent signatures is a requirement to use the node in a flow.
cluster Module¶
initialization - clustering of spikes in feature space
-
class
ClusteringNode(dtype=None)[source]¶ Bases:
botmpy.nodes.base_nodes.ResetNodeinterface for clustering algorithms
-
class
HomoscedasticClusteringNode(clus_type='kmeans', crange=[1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15], repeats=4, sigma_factor=4.0, max_iter=None, conv_thresh=None, alpha=None, cvtype='diag', gof_type='bic', dtype=None, debug=False, **kwargs)[source]¶ Bases:
botmpy.nodes.cluster.ClusteringNodeclustering with model order selection to learn a mixture model
Assuming the data are prewhitened spikes, possibly in some condensed representation e.g. PCA, the problem is to find the correct number of components and their corresponding means. The covariance matrix of all components is assumed to be the same, as the variation in the data is produced by an additive noise process. Further the covariance matrix can be assumed be the identity matrix (or a scaled version due to estimation errors, thus a spherical covariance),
To increase performance, it is assumed all necessary pre-processing has been taken care of already, to assure an optimal clustering performance (e.g.: alignment, resampling, (noise)whitening, etc.)
So we have to find the number of components and their means in a homoscedastic clustering problem. The ‘goodness of fit’ will be evaluated by evaluating a likelihood based criterion that is penalised for an increasing number of model parameters (to prevent overfitting) (ref: BIC). Minimising said criterion will lead to the most likely model.
-
execute(x, *args, **kwargs)¶ run the clustering on a set of observations
-
filter_bank Module¶
implementation of a filter bank consisting of a set of filters
-
exception
FilterBankError[source]¶ Bases:
exceptions.Exception
-
class
FilterBankNode(**kwargs)[source]¶ Bases:
mdp.Nodeabstract class that handles filter instances and their outputs
All filters constituting the filter bank have to be of the same temporal extend (Tf) and process the same channel set.
There are two different index sets. One is abbreviated “idx” and one “key”. The “idx” the index of filter in self.bank and thus a unique, hashable identifier. Where as the “key” an index in a subset of idx. Ex.: the index for list(self._idx_active_set) would be a “key”.
-
activate(idx, check=False)[source]¶ activates a filter in the filter bank
Filters are never deleted, but can be de-/reactivated and will be used respecting there activation state for the filter output of the filter bank.
No effect if idx not in self.bank.
-
ce¶
-
create_filter(xi, check=True)[source]¶ adds a new filter to the filter bank
Parameters: xi (ndarray) – template to build the filter for
-
cs¶
-
deactivate(idx, check=False)[source]¶ deactivates a filter in the filter bank
Filters are never deleted, but can be de-/reactivated and will be used respecting there activation state for the filter output of the filter bank.
No effect if idx not in self.bank.
-
execute(x)¶ Process the data contained in x.
If the object is still in the training phase, the function stop_training will be called. x is a matrix having different variables on different columns and observations on the rows.
By default, subclasses should overwrite _execute to implement their execution phase. The docstring of the _execute method overwrites this docstring.
-
filter_set¶ filter set of active filters
-
nc¶ number of channels
-
nf¶ number of filters
-
template_set¶ template set of active filters
-
tf¶ temporal filter extend [samples]
-
xcorrs¶ cross correlation tensor for active filters
-
linear_filter Module¶
filter classes for linear filters in the time domain
-
exception
FilterError[source]¶ Bases:
exceptions.Exception
-
class
FilterNode(tf, nc, ce, chan_set=None, rb_cap=None, dtype=None)[source]¶ Bases:
mdp.Nodelinear filter in the time domain
This node applies a linear filter to the data and returns the filtered data. The derivation of the filter (f) from the pattern (xi) is specified in the implementing subclass via the ‘filter_calculation’ classmethod. The template will be averaged from a ringbuffer of observations. The covariance matrix is supplied from an external covariance estimator.
-
append_xi_buf(wf, recalc=False)[source]¶ append one waveform to the xi_buffer
Parameters: - wf (ndarray) – wavefom data [self.tf, self.nc]
- recalc (bool) – if True, call self.calc_filter after appending
-
ce¶ covariance estimator
-
execute(x)¶ apply the filter to data
-
extend_xi_buf(wfs, recalc=False)[source]¶ append an iterable of waveforms to the xi_buffer
Parameters: - wfs (iterable of ndarray) – wavefom data [n][self.tf, self.nc]
- recalc (bool) – if True, call self.calc_filter after extending
-
f¶ filter (multi-channeled)
-
f_conc¶ filter (concatenated)
-
fill_xi_buf(wf, recalc=False)[source]¶ fill all of the xi_buffer with wf
Parameters: - wf : ndarrsay
ndarray of shape (self.tf, self.nc)
- recalc : bool
if True, call self.calc_filter after appending
-
classmethod
filter_calculation(xi, ce, cs, *args, **kwargs)[source]¶ ABSTRACT METHOD FOR FILTER CALCULATION
Implement this in a meaningful way in any subclass. The method should return the filter given the multichanneled template xi, the covariance estimator ce and the channel set cs plus any number of optional arguments and keywords. The filter is usually the same shape as the pattern xi.
-
nc¶ number of channels
-
plot_buffer_to_axis(axis=None, idx=None, limits=None)[source]¶ plots the current buffer on the passed axis handle
-
snr¶ signal to noise ratio (mahalanobis distance)
-
tf¶ temporal extend [sample]
-
xi¶ template (multi-channeled)
-
xi_conc¶ template (concatenated)
-
-
class
MatchedFilterNode(tf, nc, ce, chan_set=None, rb_cap=None, dtype=None)[source]¶ Bases:
botmpy.nodes.linear_filter.FilterNodematched filters in the time domain optimise the signal to noise ratio (SNR) of the matched pattern with respect to covariance matrix describing the noise background (deconvolution).
-
class
NormalisedMatchedFilterNode(tf, nc, ce, chan_set=None, rb_cap=None, dtype=None)[source]¶ Bases:
botmpy.nodes.linear_filter.FilterNodematched filters in the time domain optimise the signal to noise ratio (SNR) of the matched pattern with respect to covariance matrix describing the noise background (deconvolution). Here the deconvolution output is normalised s.t. the response of the pattern is peak of unit amplitude.
prewhiten Module¶
spike noise prewhitening algorithm
-
class
PrewhiteningNode(ncov=None, dtype=<type 'numpy.float32'>)[source]¶ Bases:
mdp.Nodeprewhitens the data with respect to a noise covariance matrix
-
execute(x, ncov=None)¶ Process the data contained in x.
If the object is still in the training phase, the function stop_training will be called. x is a matrix having different variables on different columns and observations on the rows.
By default, subclasses should overwrite _execute to implement their execution phase. The docstring of the _execute method overwrites this docstring.
-
-
class
PrewhiteningNode2(covest)[source]¶ Bases:
mdp.Nodepre-whitens data with respect to a noise covariance matrix
-
execute(x)¶ Process the data contained in x.
If the object is still in the training phase, the function stop_training will be called. x is a matrix having different variables on different columns and observations on the rows.
By default, subclasses should overwrite _execute to implement their execution phase. The docstring of the _execute method overwrites this docstring.
-
smoothing Module¶
smoothing algorithms for multichanneled data
-
class
SmoothingNode(size=5, input_dim=None, dtype=None)[source]¶ Bases:
botmpy.nodes.base_nodes.ResetNodesmooths the data using a gauss kernel of size 5 to 11
-
execute(x)¶ Process the data contained in x.
If the object is still in the training phase, the function stop_training will be called. x is a matrix having different variables on different columns and observations on the rows.
By default, subclasses should overwrite _execute to implement their execution phase. The docstring of the _execute method overwrites this docstring.
-
-
smooth(signal, window=5, kernel='gauss')[source]¶ smooth signal with kernel of type kernel and window size window
Parameters: - signal : ndarray
multichanneled signal [data, channel]
- window : ndarray
window size of the smoothing filter (len(window) < signal .shape[0])
- kernel : ndarray
- kernel to use, one of
- ‘gauss’: least squares
- ‘box’: moving average
spike_detection Module¶
detector nodes for multichanneled data
These detecors find features and feature epochs on multichanneled signals. Mostly, you will want to reset the internals of the detector after processing a chunk of data, which is featured by deriving from ResetNode. There are different kinds of detectors, distinguished by their way of feature to noise discrimination.
-
exception
EnergyNotCalculatedError[source]¶ Bases:
exceptions.ExceptionEnergyNotCalculatedError Exception
-
class
ThresholdDetectorNode(input_dim=None, output_dim=None, dtype=None, energy_func=None, threshold_func=None, threshold_mode='gt', threshold_base='energy', threshold_factor=1.0, tf=47, min_dist=1, find_max=True, ch_separate=False)[source]¶ Bases:
botmpy.nodes.base_nodes.ResetNodeabstract interface for detecting feature epochs in a signal
The ThresholdDetectorNode is the abstract interface for all detectors. The input signal is assumed to be a (multi-channeled) signal, with data for one channel in each column (or one multi-channeled observation/sample per row).
The output will be a timeseries of detected feature in the input signal. To find the features, the input signal is transformed by applying an operator (called the energy function from here on) that produces an representation of the input signal, which should optimize the SNR of the features vs the remainder of the input signal. A threshold is then applied to this energy representation of the input signal to find the feature epochs.
The output timeseries either holds the onsets of the feature epochs or the maximum of the energy function within the feature epoch, givin in samples.
Extra information about the events or the internals has to be saved in member variables along with a proper interface.
-
events¶
-
execute(x, **kwargs)¶ calls self._apply_threshold() and return the events found
-
get_epochs(cut=None, invert=False, merge=False)[source]¶ returns epochs based on self.events for the current iteration
Parameters: - cut : (int,int)
Window size of an epoch in samples (befor,after) the event sample. If None, self._tf will be used.
- invert : bool
Inverts the epochs, frex to yield noise epochs instead of spike epochs.
- merge : bool
Merges overlapping epochs.
Returns: - ndarray
ndarray with epochs on the rows [[start,end]]
-
get_extracted_events(mc=False, align_at=-1, kind='min', rsf=1.0, buffer=False)[source]¶ yields the extracted spikes
Parameters: - mc (bool) – if True, return multichannel events, else return concatenated events. Default=False
- align_at (int or float) – if a float from (0.0,1.0), determine the align_sample according to that weight. If a positive integer from (0, self.tf-1] use that sample as the align_sample. Default=0.25 * self.tf
- kind (str) – one of “min”, “max”, “energy” or “none”. method to use for alignment, will be passed to the alignment function. Default=’min’
- rsf (float) – resampling factor (use integer values of powers of 2)
- buffer (bool) – if True, write to buffer regardless of current buffer state. Default=False
-
stop_training(*args, **kwargs)¶ Stop the training phase.
By default, subclasses should overwrite _stop_training to implement this functionality. The docstring of the _stop_training method overwrites this docstring.
-
train(x)¶ Update the internal structures according to the input data x.
x is a matrix having different variables on different columns and observations on the rows.
By default, subclasses should overwrite _train to implement their training phase. The docstring of the _train method overwrites this docstring.
Note: a subclass supporting multiple training phases should implement the same signature for all the training phases and document the meaning of the arguments in the _train method doc-string. Having consistent signatures is a requirement to use the node in a flow.
-
-
class
SDAbsNode(**kwargs)[source]¶ Bases:
botmpy.nodes.spike_detection.ThresholdDetectorNodespike detector
energy: absolute of the signal threshold: signal.std
-
class
SDSqrNode(**kwargs)[source]¶ Bases:
botmpy.nodes.spike_detection.ThresholdDetectorNodespike detector
energy: square of the signal threshold: signal.var
-
class
SDMteoNode(kvalues=[1, 3, 5, 7, 9], quantile=0.98, **kwargs)[source]¶ Bases:
botmpy.nodes.spike_detection.ThresholdDetectorNodespike detector
energy: multiresolution teager energy operator threshold: energy.std
-
class
SDKteoNode(kvalue=1, quantile=0.98, **kwargs)[source]¶ Bases:
botmpy.nodes.spike_detection.ThresholdDetectorNodespike detector
energy: teager energy operator threshold: energy.std
-
class
SDIntraNode(**kwargs)[source]¶ Bases:
botmpy.nodes.spike_detection.ThresholdDetectorNodespike detector
energy: identity threshold: zero
-
class
SDPeakNode(**kwargs)[source]¶ Bases:
botmpy.nodes.spike_detection.ThresholdDetectorNodespike detector
energy: absolute of the signal threshold: signal.std
spike_sorting Module¶
implementation of spike sorting with matched filters
See: [1] F. Franke, M. Natora, C. Boucsein, M. Munk, and K. Obermayer. An online spike detection and spike classification algorithm capable of instantaneous resolution of overlapping spikes. Journal of Computational Neuroscience, 2009 [2] F. Franke, ... , 2012, The revolutionary BOTM Paper
-
class
FilterBankSortingNode(**kwargs)[source]¶ Bases:
botmpy.nodes.filter_bank.FilterBankNodeabstract class that handles filter instances and their outputs
This class provides a pipeline structure to implement spike sorting algorithms that operate on a filter bank. The implementation is done by implementing the self._pre_filter, self._post_filter, self._pre_sort, self._sort_chunk and self._post_sort methods with meaning full processing. After the filter steps the filter output is present and can be processed on. Input data can be partitioned into chunks of smaller size.
-
execute(x)¶ Process the data contained in x.
If the object is still in the training phase, the function stop_training will be called. x is a matrix having different variables on different columns and observations on the rows.
By default, subclasses should overwrite _execute to implement their execution phase. The docstring of the _execute method overwrites this docstring.
-
plot_sorting(ph=None, show=False)[source]¶ plot the sorting of the last data chunk
Parameters: - ph (plot handle) – plot handle top use for the plot
- show (bool) – if True, call plt.show()
-
plot_sorting_waveforms(ph=None, show=False, **kwargs)[source]¶ plot the waveforms of the sorting of the last data chunk
Parameters: - ph (plot handle) – plot handle to use for the
- show (bool) – if True, call plt.show()
-
spikes_u(u, mc=True, exclude_overlaps=True, overlap_window=None, align_at=-1, align_kind='min', align_rsf=1.0)[source]¶ yields the spike for the u-th filter
Parameters: - u (int) – index of the filter # CHECK THIS
- mc (bool) – if True, return spikes multi-channeled, else return spikes concatenated Default=True
- exclude_overlaps (bool) – if True, exclude overlap spike
- overlap_window (int) – if exclude_overlaps is True, this will define the overlap range, if None set overlap_window=self._tf. Default=None
-
-
class
AdaptiveBayesOptimalTemplateMatchingNode(**kwargs)[source]¶ Bases:
botmpy.nodes.spike_sorting.BayesOptimalTemplateMatchingNodeAdaptive BOTM Node
adaptivity here means,backwards sense, that known templates and covariances are adapted local temporal changes. In the forward sense a parallel spike detection is matched to find currently unidenified units in the data.
-
det¶
-
execute(x, ex_st=None)¶ Process the data contained in x.
If the object is still in the training phase, the function stop_training will be called. x is a matrix having different variables on different columns and observations on the rows.
By default, subclasses should overwrite _execute to implement their execution phase. The docstring of the _execute method overwrites this docstring.
-
-
class
BayesOptimalTemplateMatchingNode(**kwargs)[source]¶ Bases:
botmpy.nodes.spike_sorting.FilterBankSortingNodeFilterBanksSortingNode derivative for the BOTM algorithm
Can use two implementations of the Bayes Optimal Template-Matching (BOTM) algorithm as presented in [2]. First implementation uses explicitly constructed overlap channels for the extend of the complete input signal, the other implementation uses subtractive interference cancellation (SIC) on epochs of the signal, where the template discriminants are greater the the noise discriminant.
-
component_divergence(obs, with_noise=False, loading=False, subdim=None)[source]¶ component probabilities under the model
Parameters: - obs (ndarray) – observations to be evaluated [n, tf, nc]
- with_noise (bool) – if True, include the noise cluster as component in the mixture. Default=False
- loading (bool) – if True, use the loaded matrix Default=False
- subdim (int) – dimensionality of subspace to build the inverse over. if None ignore Default=None
Return type: ndarray
Returns: divergence from means of current filter bank[n, c]
-
copy= <module 'copy' from '/home/docs/.pyenv/versions/2.7.13/lib/python2.7/copy.pyc'>¶
-
noise_prior¶
-
posterior_prob(obs, with_noise=False)[source]¶ posterior probabilities for data under the model
Parameters: - obs (ndarray) – observations to be evaluated [n, tf, nc]
- with_noise (bool) – if True, include the noise cluster as component in the mixture. Default=False
Return type: ndarray
Returns: matrix with per component posterior probabilities [n, c]
-
spike_prior¶
-
spike_prior_bias¶
-
-
BOTMNode¶ alias of
BayesOptimalTemplateMatchingNode
-
ABOTMNode¶